Annotations Import
Darwin fundamentals
Darwin advanced
Play video
7:22

Play video
7:22

Play video
7:22
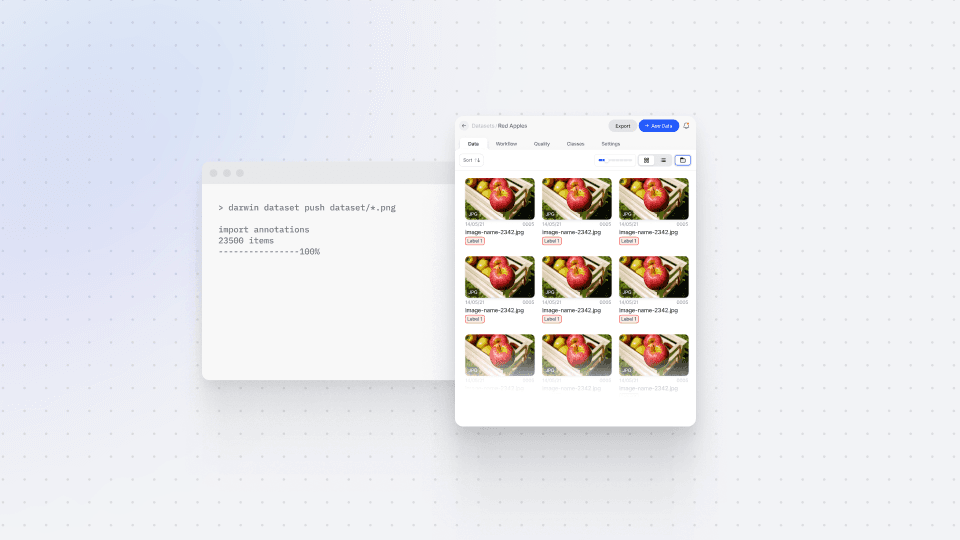
In this Darwin Advanced session, we discuss how you can import annotations to V7 by using the command line interface (CLI) or the software development kit (SDK) - in any of our supported file formats. Head to our Import Annotations documentation to access a detailed overview of the process and the scripts discussed in the video.
This tutorial begins with a breakdown of the Darwin JSON file format, the most versatile and widely used option for importing annotations. However, if you require another format - don’t worry, we support COCO, YOLO, Instances PNG, CVAT, and many others.
We begin by breaking down the makeup of the Darwin JSON format, highlighting the annotation and item data specific to the image or video you’re importing. To get started with annotations import, we showcase two methods: the CLI and the SDK.
The step-by-step run-through of the CLI method includes how users can authenticate themselves using the “darwin authenticate” command, and how they can access datasets by using “darwin datasets remote”.
Next, we demonstrate the process of using the SDK, with code examples. Here we walk you through initiating the client and specifying the annotation format, to uploading the annotations to the dataset hosted on V7.
You’ll come away with a detailed and accessible explanation of how to import annotations to V7, allowing you to organize and drive your computer vision projects forward.
In this Darwin Advanced session, we discuss how you can import annotations to V7 by using the command line interface (CLI) or the software development kit (SDK) - in any of our supported file formats. Head to our Import Annotations documentation to access a detailed overview of the process and the scripts discussed in the video.
This tutorial begins with a breakdown of the Darwin JSON file format, the most versatile and widely used option for importing annotations. However, if you require another format - don’t worry, we support COCO, YOLO, Instances PNG, CVAT, and many others.
We begin by breaking down the makeup of the Darwin JSON format, highlighting the annotation and item data specific to the image or video you’re importing. To get started with annotations import, we showcase two methods: the CLI and the SDK.
The step-by-step run-through of the CLI method includes how users can authenticate themselves using the “darwin authenticate” command, and how they can access datasets by using “darwin datasets remote”.
Next, we demonstrate the process of using the SDK, with code examples. Here we walk you through initiating the client and specifying the annotation format, to uploading the annotations to the dataset hosted on V7.
You’ll come away with a detailed and accessible explanation of how to import annotations to V7, allowing you to organize and drive your computer vision projects forward.
In this Darwin Advanced session, we discuss how you can import annotations to V7 by using the command line interface (CLI) or the software development kit (SDK) - in any of our supported file formats. Head to our Import Annotations documentation to access a detailed overview of the process and the scripts discussed in the video.
This tutorial begins with a breakdown of the Darwin JSON file format, the most versatile and widely used option for importing annotations. However, if you require another format - don’t worry, we support COCO, YOLO, Instances PNG, CVAT, and many others.
We begin by breaking down the makeup of the Darwin JSON format, highlighting the annotation and item data specific to the image or video you’re importing. To get started with annotations import, we showcase two methods: the CLI and the SDK.
The step-by-step run-through of the CLI method includes how users can authenticate themselves using the “darwin authenticate” command, and how they can access datasets by using “darwin datasets remote”.
Next, we demonstrate the process of using the SDK, with code examples. Here we walk you through initiating the client and specifying the annotation format, to uploading the annotations to the dataset hosted on V7.
You’ll come away with a detailed and accessible explanation of how to import annotations to V7, allowing you to organize and drive your computer vision projects forward.
Up next
3:49
Watch video
Auto-Annotate Tool
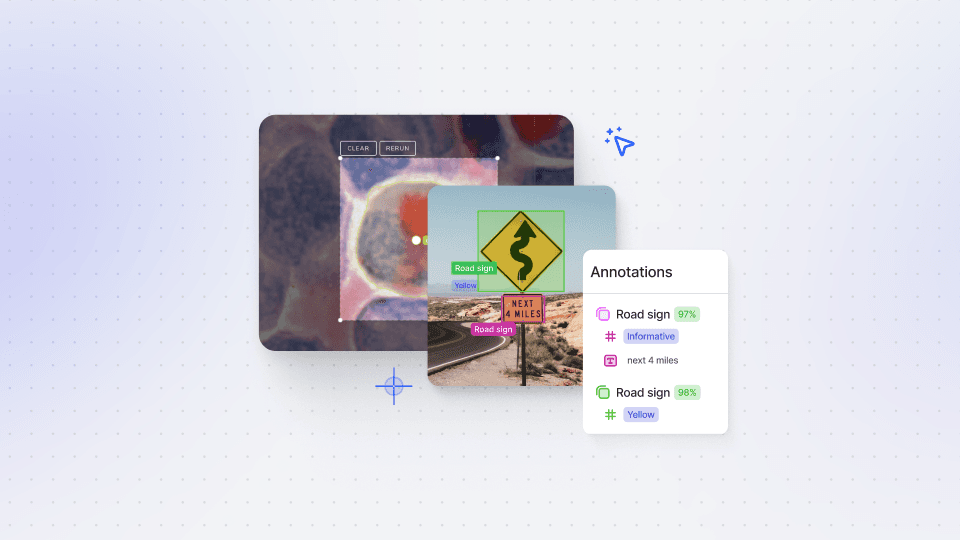
How does the Auto-Annotate tool work? We tackle accurate polygon and pixel-wise annotation masks.
3:49
Watch video

Auto-Annotate Tool
How does the Auto-Annotate tool work? We tackle accurate polygon and pixel-wise annotation masks.
3:49
Watch video

Auto-Annotate Tool
How does the Auto-Annotate tool work? We tackle accurate polygon and pixel-wise annotation masks.
16:35
Watch video
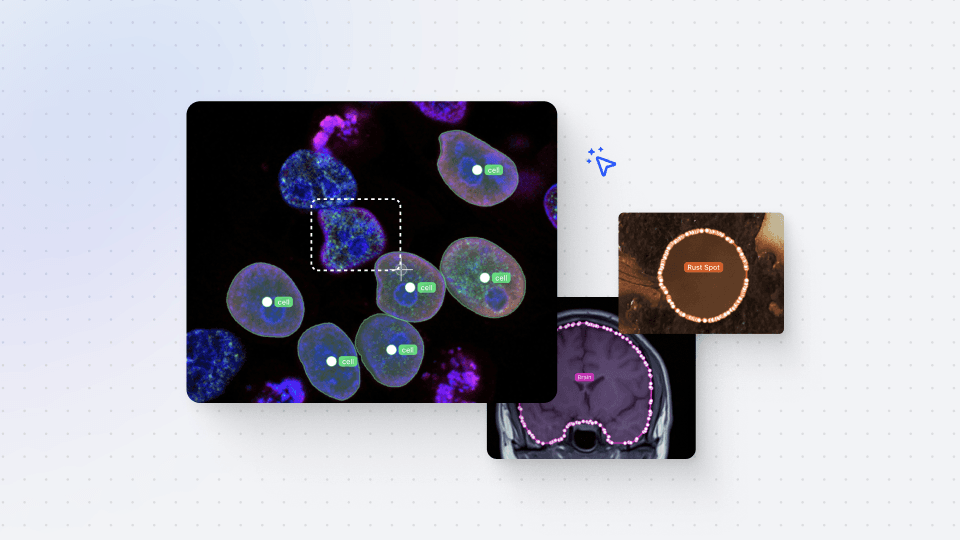
Annotations - Getting Started
We dive into image annotation and explain how you can get the most out of these impressive features.
16:35
Watch video

Annotations - Getting Started
We dive into image annotation and explain how you can get the most out of these impressive features.
16:35
Watch video

Annotations - Getting Started
We dive into image annotation and explain how you can get the most out of these impressive features.
5:56
Watch video

Auto-Annotate Tips & Tricks
Wield Auto-Annotate like the experts, with tips and tricks to accelerate your use of the tool.
5:56
Watch video

Auto-Annotate Tips & Tricks
Wield Auto-Annotate like the experts, with tips and tricks to accelerate your use of the tool.
5:56
Watch video

Auto-Annotate Tips & Tricks
Wield Auto-Annotate like the experts, with tips and tricks to accelerate your use of the tool.