Annotation suite
Data labeling for Frontier Models.
Lightning-fast, expert-driven.
Data labeling for Frontier Models.
Lightning-fast, expert-driven.
Customers
Build breakthrough AI models.
Join leading AI research labs and Fortune 100s.
Customers
Annotation suite
Create ground truth 10x faster without errors.
Manage expert labelers with AI-assisted annotation and review.
Demo
See it in action
Build in a visual, no-code environment
See it in action
Build in a visual, no-code environment
See it in action
Build in a visual, no-code environment

Dataset
Consensus
Logic
Complete
Dataset
5



Rail Maintenance
6
Files
4
Completed
2
In progress
66.67%
darwin.v7labs.com
Dataset
Consensus
Logic
Complete
Dataset
5



Rail Maintenance
6
Files
4
Completed
2
In progress
66.67%
Suited for every industry.
Create your custom workflow.
RLHF + RLAIF
RLHF + RLAIF
RLHF + RLAIF
Dataset
Dataset
Review
Review
Logic
Logic
Annotate
Annotate
Complete
Complete
Model-in-the-loop
Model-in-the-loop
Model-in-the-loop
Dataset
Dataset
Model
Model
Annotate
Annotate
Review
Review
Complete
Complete
Webhook
Webhook
Webhook
Dataset
Dataset
Annotate
Annotate
Review
Review
Webhook
Webhook

"Visibility on metrics in V7 is very helpful to us, and it's something we didn’t have in our internal solution."
Andrew Achkar
Technical Director at Miovison
"Visibility on metrics in V7 is very helpful to us, and it's something we didn’t have in our internal solution."

Andrew Achkar
Technical Director at Miovison
Automated labeling
Annotate any dataset
with speed and accuracy
Annotate any dataset
with speed and accuracy
Annotate any dataset
with speed and accuracy
For many AI teams, creating high-quality training datasets is their biggest bottleneck. Annotation projects often stretch over months, consuming thousands of hours of meticulous work. V7 can speed up data annotation 10x, turning a months-long process into weeks.
Use intuitive interface and AI-assisted tools to turn complex labeling tasks into a few simple clicks and adjustments. Work with any size dataset and file type, from videos, PDFs, and architectural drawings to specialized medical formats like SVS or DICOM.


AI-assisted data labeling
Label data at lightning speed with V7 Auto-Annotate and SAM2. Segment complex objects like lesions in CT scans and items on assembly lines with high accuracy. Achieve expert level segmentation across diverse domains, regardless of industry.
AI-assisted data labeling
Label data at lightning speed with V7 Auto-Annotate and SAM2. Segment complex objects like lesions in CT scans and items on assembly lines with high accuracy. Achieve expert level segmentation across diverse domains, regardless of industry.
Auto-track for video
Track objects across selected time ranges in videos. Automatically follow instances and mark in-and-out of view situations. Streamline video annotation for tasks like AI-assisted surgeries, retail shrinkage prevention, or sports analytics.
Auto-track for video
Track objects across selected time ranges in videos. Automatically follow instances and mark in-and-out of view situations. Streamline video annotation for tasks like AI-assisted surgeries, retail shrinkage prevention, or sports analytics.
Label similar objects
Pick one object and find similar ones automatically. Speed up repetitive annotation problems like preparing training data for cell counting models or product identification on shelves. Reduce manual effort in large-scale labeling projects. Reduce human errors caused by fatigue and monotony.
Label similar objects
Pick one object and find similar ones automatically. Speed up repetitive annotation problems like preparing training data for cell counting models or product identification on shelves. Reduce manual effort in large-scale labeling projects. Reduce human errors caused by fatigue and monotony.

Model in the loop
Integrate external or out-of-the-box models to pre-label your data or detect quality issues. Compare performance between human labelers and AI models in blind tests. Connect models to your ML pipeline and improve your annotation process and model accuracy.

Model in the loop
Integrate external or out-of-the-box models to pre-label your data or detect quality issues. Compare performance between human labelers and AI models in blind tests. Connect models to your ML pipeline and improve your annotation process and model accuracy.
Use cases
Create high-quality training data.
V7 supports all your labeling needs.
Images
Videos
DICOM & NIfTI
Microscopy
RLHF
Speed up complex annotation tasks with AI-assisted labeling and custom computer vision models. Generate semantic pixel masks and segment irregular shapes with a single click. Use keypoints, brushes, polylines, and more.
Images
Videos
DICOM & NIfTI
Microscopy
RLHF
Speed up complex annotation tasks with AI-assisted labeling and custom computer vision models. Generate semantic pixel masks and segment irregular shapes with a single click. Use keypoints, brushes, polylines, and more.
In numbers
Faster annotation
Faster annotation
Faster annotation
Data types supported
Data types supported
Data types supported
Platform Uptime
Platform Uptime
Platform Uptime
Workflows
Leverage humans in the loop.
Build better ML pipelines.
Leverage humans in the loop. Build better ML pipelines.
Leverage humans in the loop.
Build better ML pipelines.
As annotation projects scale, AI teams struggle to track progress, maintain QA standards, and ensure accountability. Inconsistencies can quickly compound, leading to compromised data quality—which impacts the entire AI development lifecycle.
V7 allows you to design multi-stage review workflows to orchestrate your labeling process. Assign roles, tasks, and manage project completion. Use conditional logic and automations to always route data to the right stages or team members.
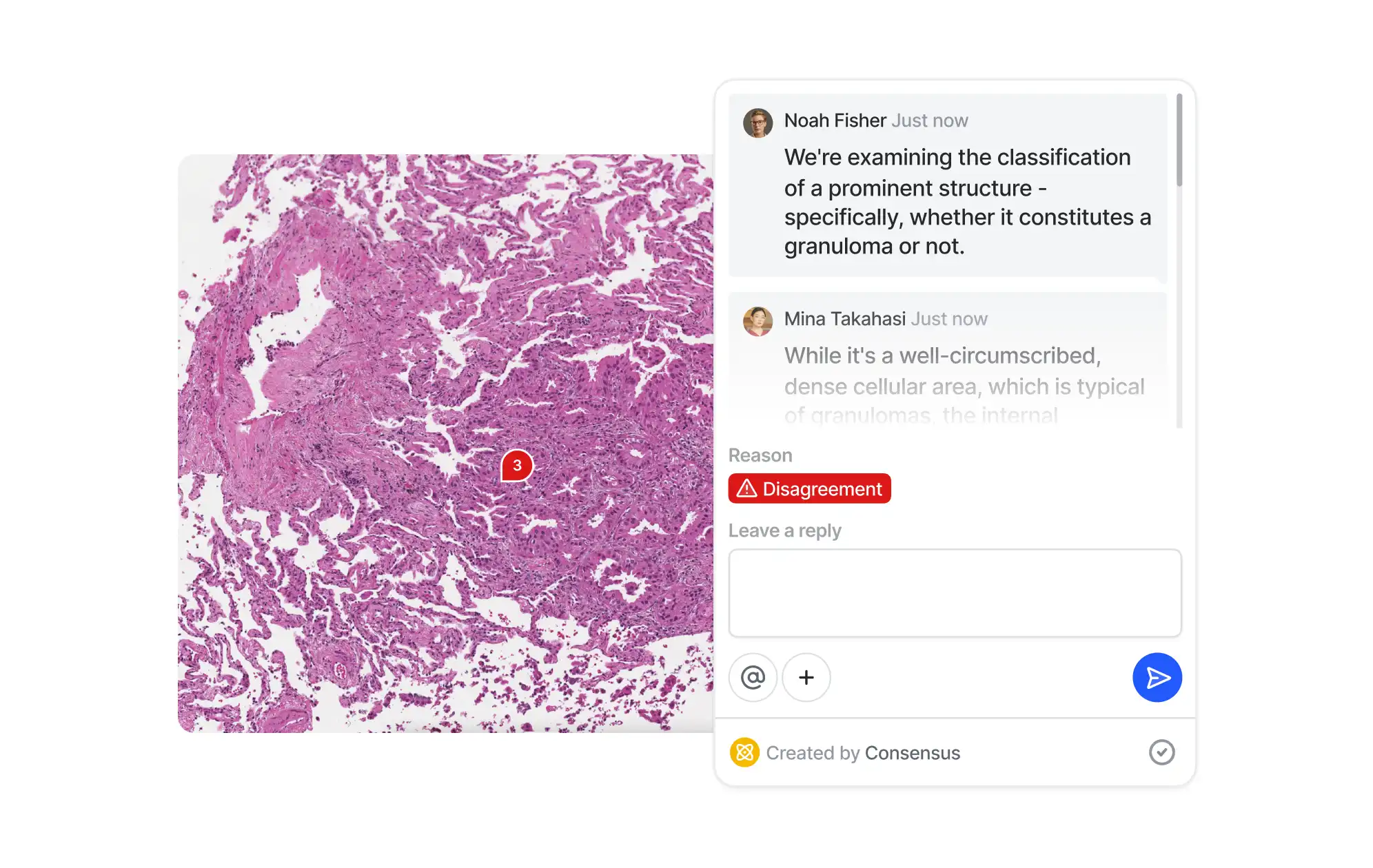
Collaborate
Improve teamwork with real-time collaboration features. Ensure smooth communication between annotators, reviewers, and ML engineers to speed up assignments. Delegate tasks and track progress of multiple independent annotation projects.
Consensus
Evaluate labelers
Data labeling services
Collaborate
Improve teamwork with real-time collaboration features. Ensure smooth communication between annotators, reviewers, and ML engineers to speed up assignments. Delegate tasks and track progress of multiple independent annotation projects.
Consensus
Evaluate labelers
Data labeling services

Labeling services
Specialized data annotation
for modern AI challenges
Build vs Buy
Testimonials
Secure and trustworthy AI.
Here's what our customers say.
Secure and trustworthy AI.
Here's what our customers say.
Secure and trustworthy AI.
Here's what our customers say.
Security & Integrations
Connect your data with confidence.
Built for ML developers.
Connect your data with confidence. Built for ML developers.
Connect your data with confidence.
Built for ML developers.
Security & Integrations
Integrate with your toolstack
Integrate V7 with your existing ML infrastructure. Use your private cloud with major providers including AWS, Google Cloud, and Azure. Export annotations in formats compatible with popular frameworks like TensorFlow and PyTorch.
Integrate with your toolstack
Integrate V7 with your existing ML infrastructure. Use your private cloud with major providers including AWS, Google Cloud, and Azure. Export annotations in formats compatible with popular frameworks like TensorFlow and PyTorch.
Built with security in mind

Benefit from SOC 2 Type II and HIPAA-compliant infrastructure. Set granular access controls and track all actions with detailed logs. Meet stringent security standards for industries like healthcare and finance.
Built with security in mind

Benefit from SOC 2 Type II and HIPAA-compliant infrastructure. Set granular access controls and track all actions with detailed logs. Meet stringent security standards for industries like healthcare and finance.
Developer friendly API & SDK

V7 is built by developers for developers. Our tools make it easy to automate workflows, customize integrations, and extend V7's capabilities to fit your unique use cases. Enjoy clear documentation, code samples, and responsive support.
Developer friendly API & SDK

V7 is built by developers for developers. Our tools make it easy to automate workflows, customize integrations, and extend V7's capabilities to fit your unique use cases. Enjoy clear documentation, code samples, and responsive support.
Built for scale

Handle projects of any size. Create multiple workspaces and invite users to collaborate. Manage and filter datasets with intuitive GUI dashboards. Set up custom views, add tags, and save advanced filtering conditions.
Built for scale

Handle projects of any size. Create multiple workspaces and invite users to collaborate. Manage and filter datasets with intuitive GUI dashboards. Set up custom views, add tags, and save advanced filtering conditions.
Next steps
Tell us more about your project.
Let's build a proof of concept.
Try our free tier or talk to one of our experts.



Next steps
Tell us more about your project.
Let's build a proof of concept.






























